Happy to announce that our paper about Linear-combination modeling of GABA-edited MRS is now available at NMR in Biomedicine. A well-parameterized macromolecule (MM) basis function at 3 ppm improved overall modeling performance.
GABA-edited MRS is best quantified by LCM with a well-parameterized coedited MM basis function constraint to the non-overlapped macromolecule peak at 0.9 ppm combined with sparse spline knot spacing and modeling range between 0.5 and 4 ppm.
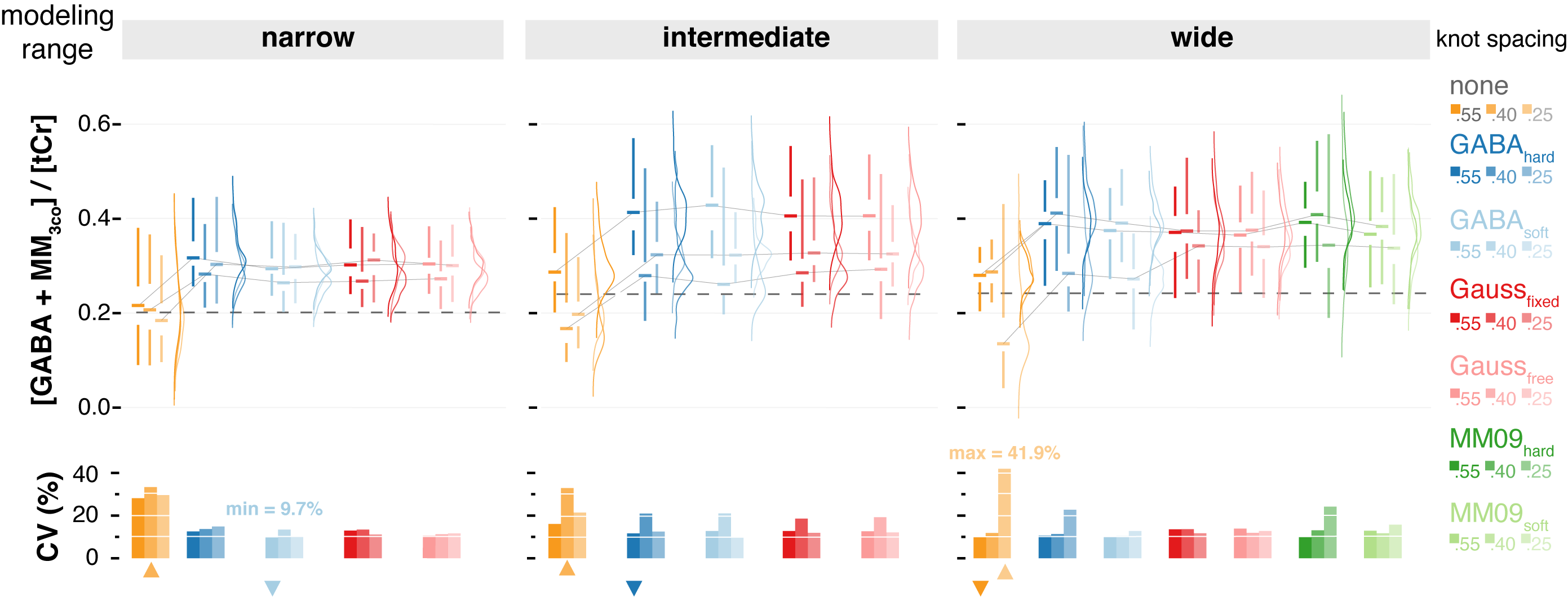 Graphical abstract: Smallest GABA+ CVs were found when the co-edited MM was hard constraint by the non-overlapped 0.9 ppm macromolecule peak.
Graphical abstract: Smallest GABA+ CVs were found when the co-edited MM was hard constraint by the non-overlapped 0.9 ppm macromolecule peak.
Thank you to those who contributed to this work - co-authors, my mentors and supervisors Georg Oeltzschner and Richard Edden.



