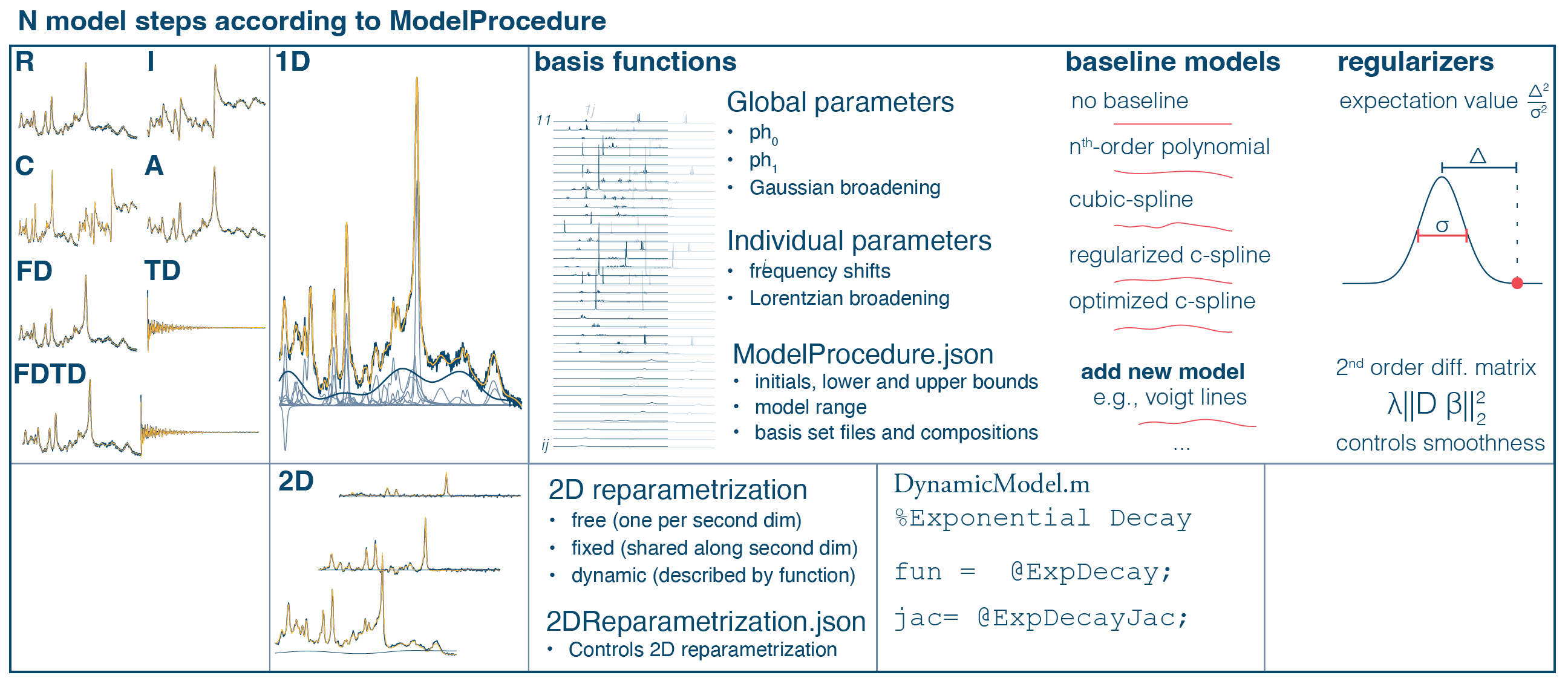
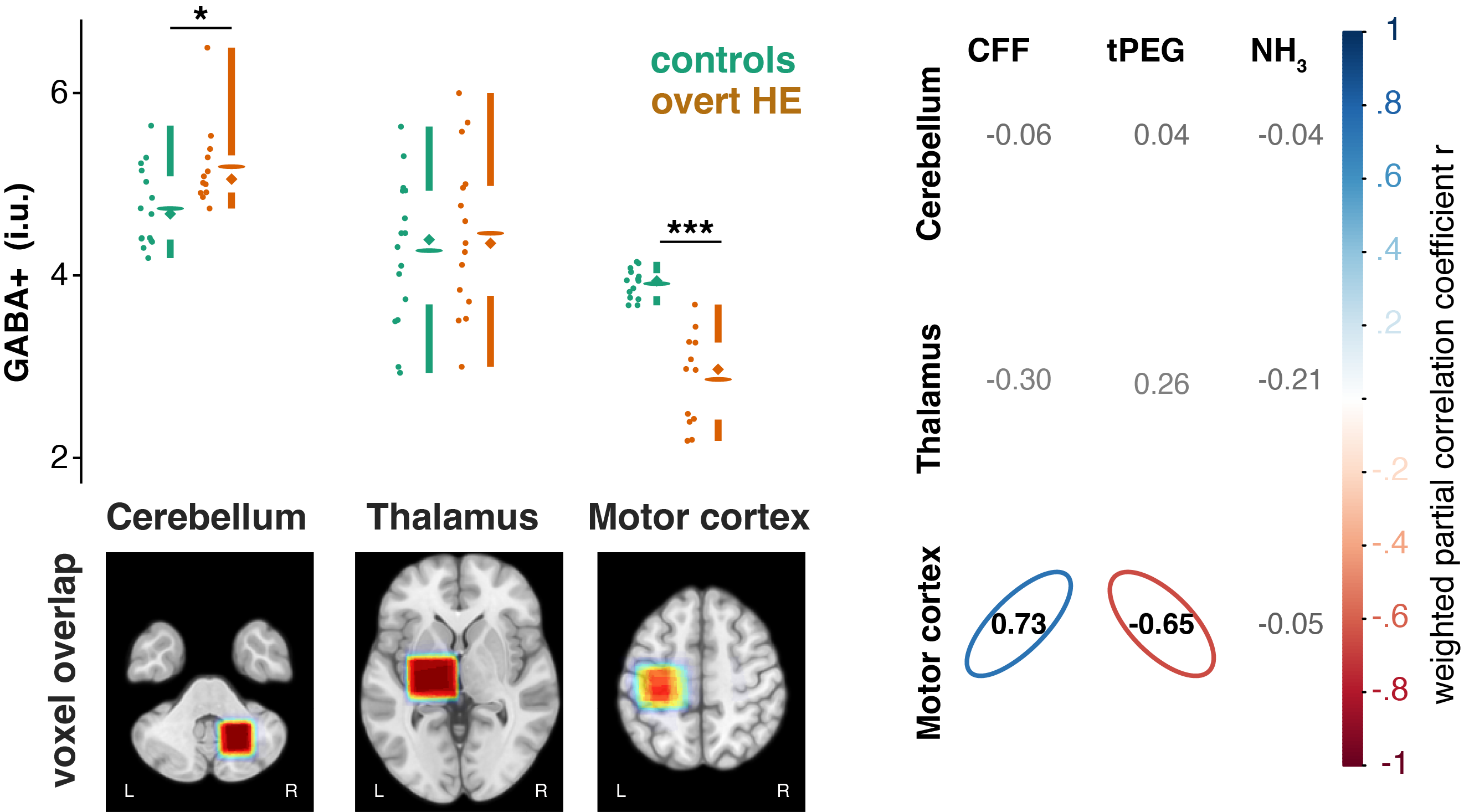
Continuous automated MRS data analysis workflow for applied studies
Are you afraid of loosing your MRS data during a MRS application study? Check out our new preprint on bioRxiv. Here we present a continuous automated MRS analysis workflow. This workflow integrates automated data sorting, conversion to NIfTI-MRS, analysis in Osprey, and automated reproting of the results.
[Read More]